Tag: assembler
-
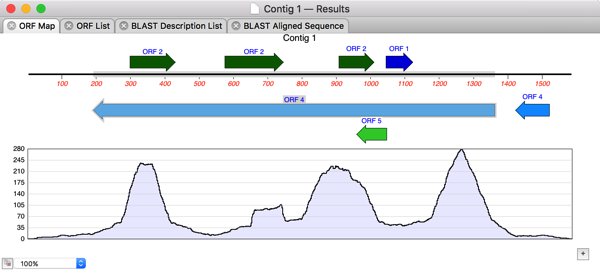
MacVectorTip: Assembling Nanopore or PacBio Long-Read Data with Flye
In the previous post we discussed the various ways in which you can analyze Oxford Nanopore’s long read data. For de novo assembly we recommend using Flye, which can also be used with PacBio data. Here are some tips to get the most out of Flye. IMPORTANT: MacVector simply wrappers around the Flye executable algorithm which depends on…
-
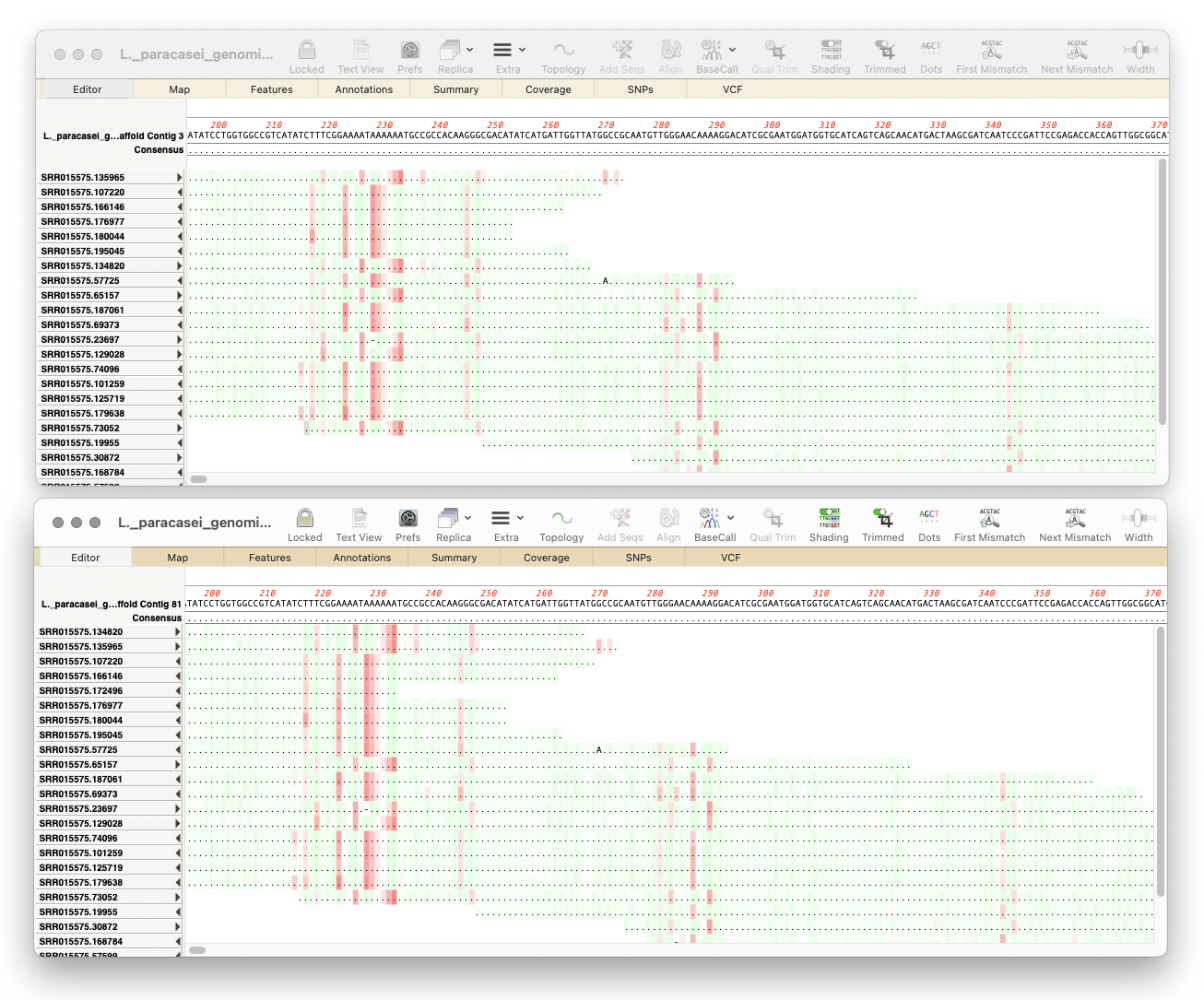
MacVectorTip: Working with Oxford Nanopore Long Read Data
Here’s a few tips regarding analyzing long read data from the Oxford Nanopore Technology MinION and GridION machines. First, you should always first create a File | New | Assembly Project and then (typically) click on the Add Reads toolbar button and select the appropriate fastq formatted data files. These are often supplied in compressed…
-
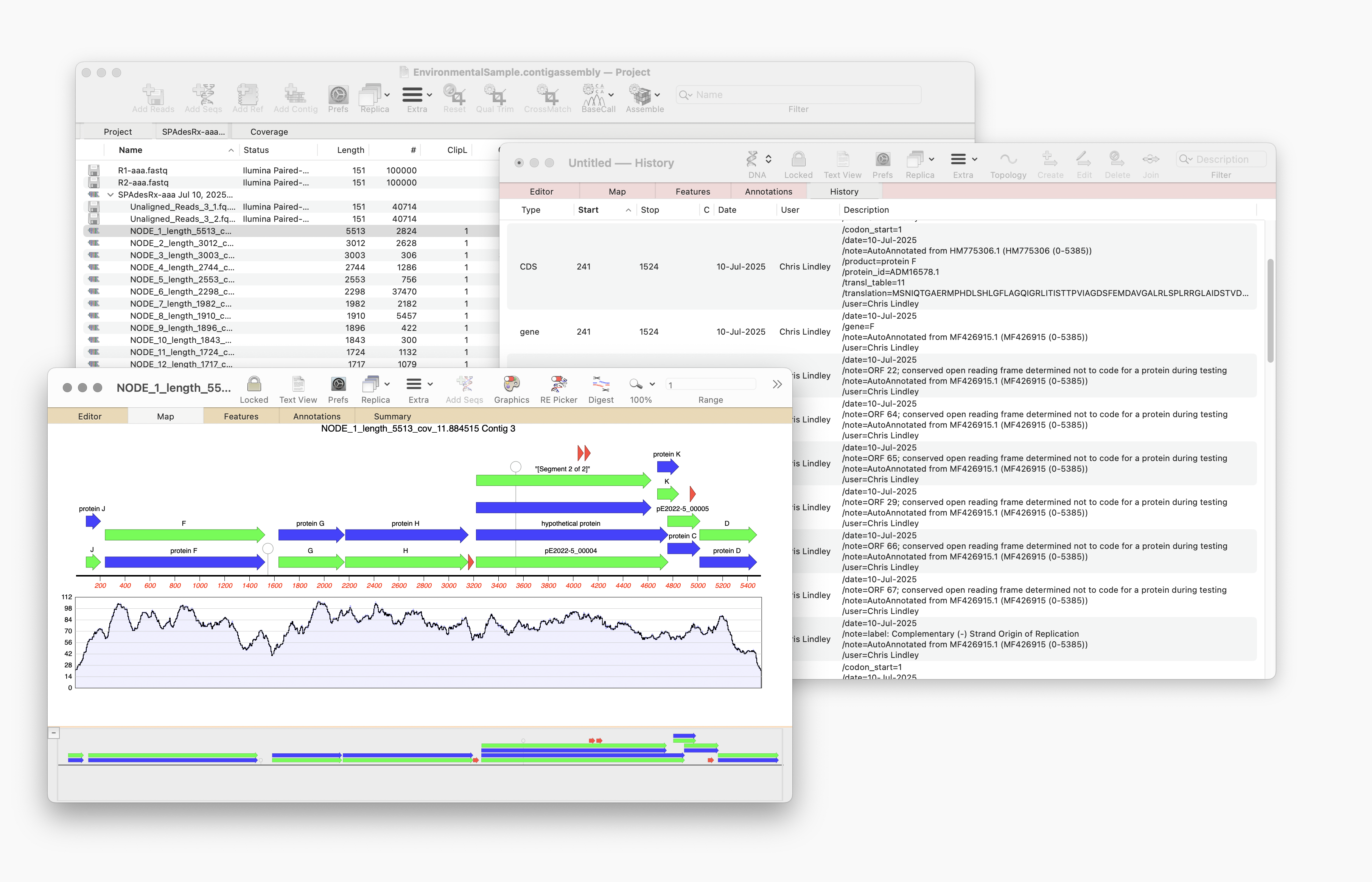
Automating annotation of sequences via BLAST
MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Ideal for identifying contigs from a de novo assembly. One of these new tools is AutoAnnotate (via BLAST) Auto-Annotate (via BLAST) is similar to Auto-Annotate (local), except instead of using curated sequences on your own…
-
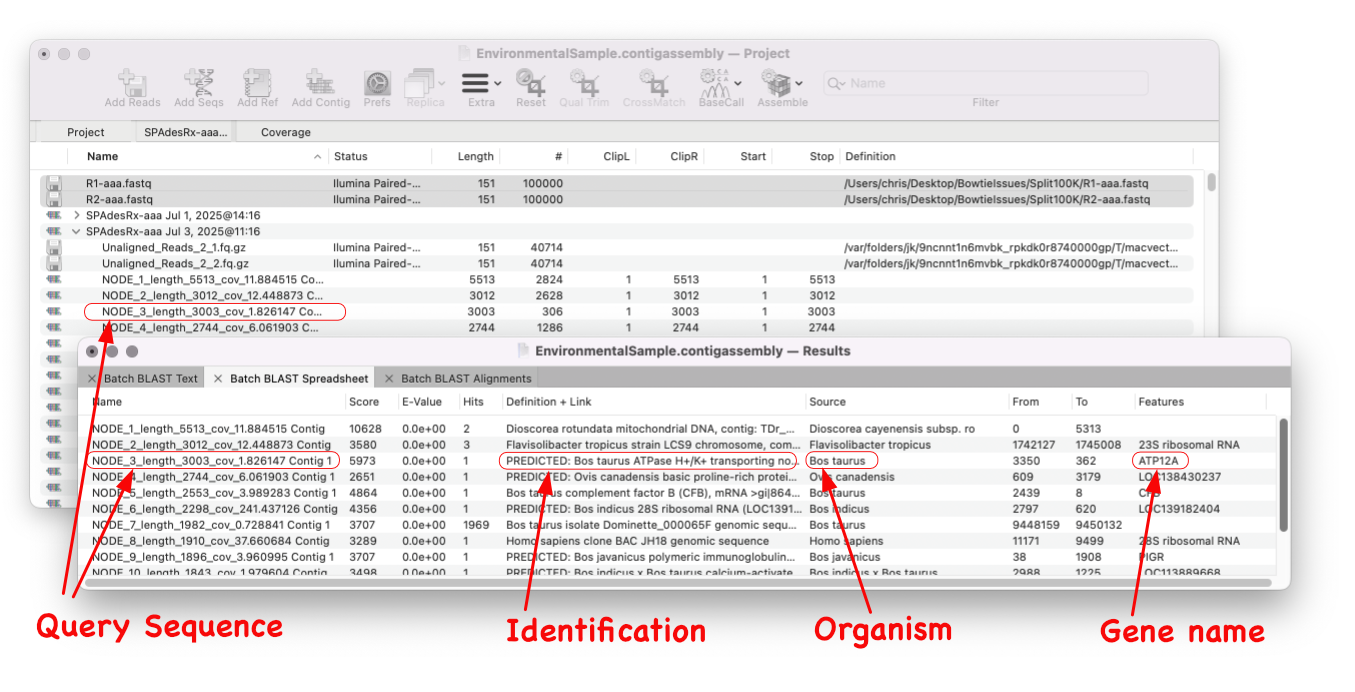
MacVector 18.8 Is Here – Packed with Powerful New Features
A game changer for identifying unknown sequences MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Batch BLAST allows you to automate BLAST searches for multiple sequences and Auto-Annotate (via BLAST) allows you to automatically annotate multiple sequences.…
-
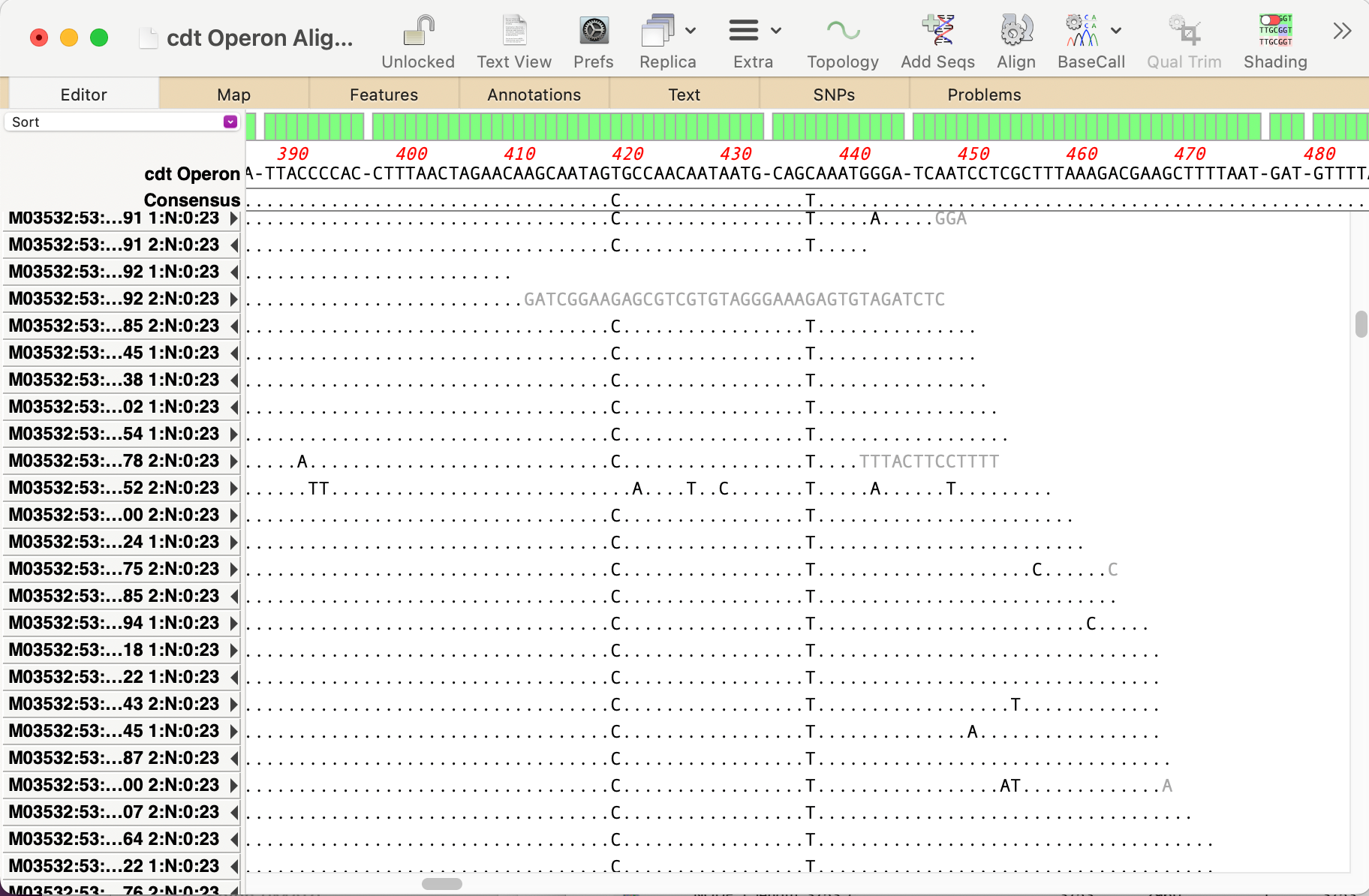
MacVectorTip: Use Align to Folder to filter NGS data for specific genes
Even the latest Macintosh computers loaded with as much RAM as you can afford will still struggle to de novo assemble genomes much over 50 Mbp. But, often that is not required. If you are just interested in a few genes, or a specific region of a chromosome, you can use Align to Folder to filter the…
-
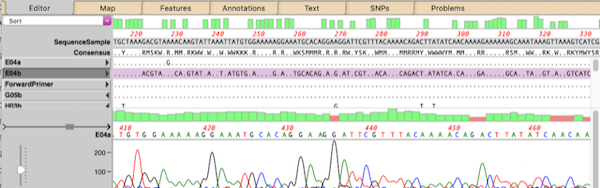
MacVector Pro is now MacVector Pro with Assembler
MacVector Pro now includes Assembler, a powerful sequence assembly plugin that brings sequence assembly directly to your desktop with the same user-friendly interface MacVector users have come to expect. Assembler simplifies the management, assembly, and analysis of all types of sequencing data. Assembler’s extensive toolkit has always been seamlessly integrated into MacVector, but previously required…
-

MacVector 18.7: Long-Read Reference Alignments using minimap2
Our latest release, MacVector 18.7, sees the addition of Minimap2 to Assembler’s sequencing toolkit. So if you have the Assembler module, you can now map noisy long-read data from Pacific Biosciences or Oxford Nanopore to one or more genomes. Minimap2 is a reference assembler similar to Bowtie2. But whereas Bowtie2 excels at mapping “short reads” (500nt or less) to…
-

MacVector 18.7 has just been released.
MacVector 18.7 has just been released. If you are eligible for this release you will be prompted to upgrade, otherwise go to MACVECTOR | CHECK FOR UPDATES… and follow the prompts to be automatically upgraded. If your license is not eligible then why not upgrade? Overview MacVector 18.7 introduces a History tab to track the…
-
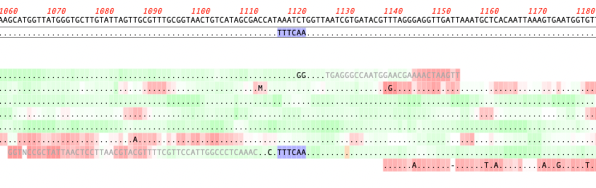
MacVectorTip: Quality scoring of manual edits to your contigs.
Quality scoring of Assemblies and Align to Reference alignments can be visualized directly on the sequence. Residues can be shaded according to their quality scores. These can be displayed anywhere quality values are available, including de novo and reference assemblies in Assembler and Align to Reference alignments. The intensity of the shading of residues indicates…
-
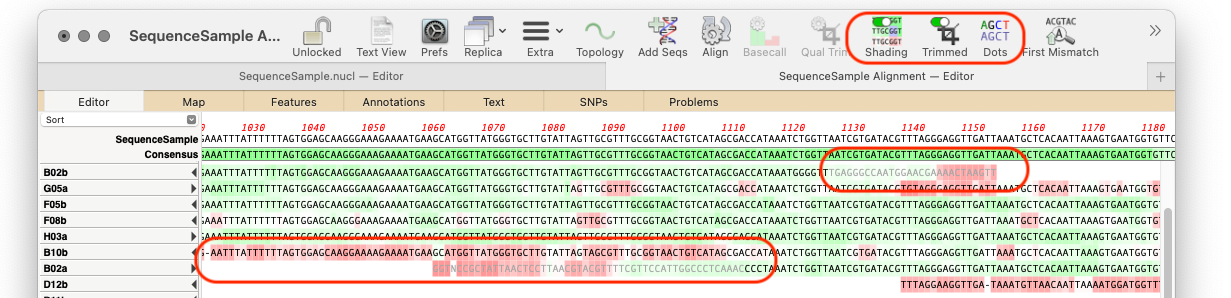
MacVectorTip: Trimming by Quality in sequence assemblies
Many of our users may be familiar with the ability of Sequencher to semi-automatically trim poor quality sequences from the ends of Sanger ABI reads. Although it is generally not necessary to do this in MacVector because most of the algorithms can automatically handle poor quality data, there are times when it can be beneficial.…
