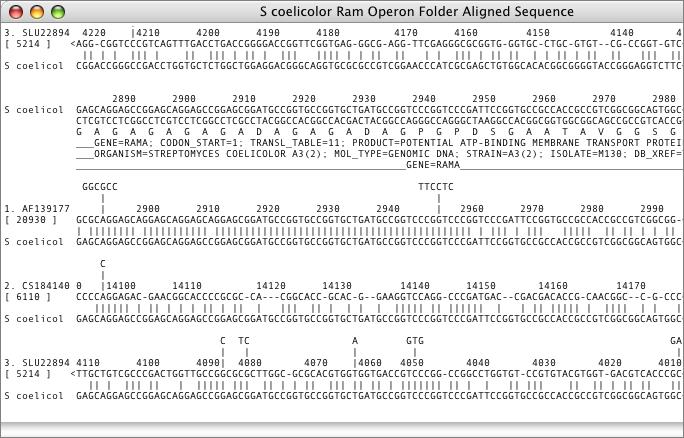
NCBI Entrez
The National Center for Biotechnology Information (NCBI) maintains a number of sequence databases that can be accessed over the Internet. MacVector has a built-in Internet database browser that lets you search the NCBI’s Entrez databases for sequences using combinations of search terms such as author, organism, keywords, accession number etc.
Sequences that match the search terms can be downloaded either individually or in batches to your desktop or to a folder on your hard drive. Sequences are retrieved with all of their associated annotations and features intact. These can be viewed and edited in MacVector, or used for additional analyses.
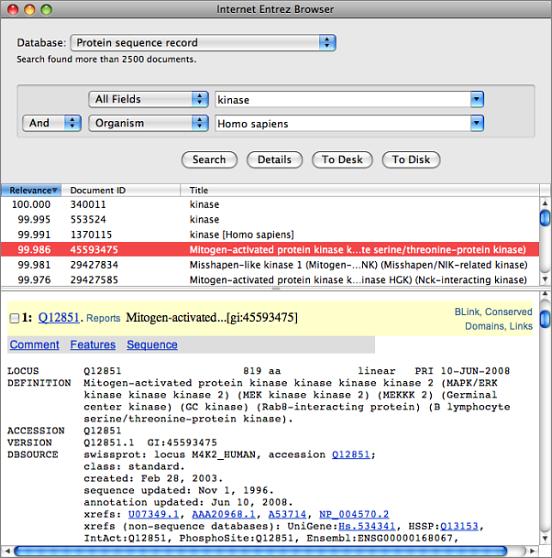
NCBI BLAST
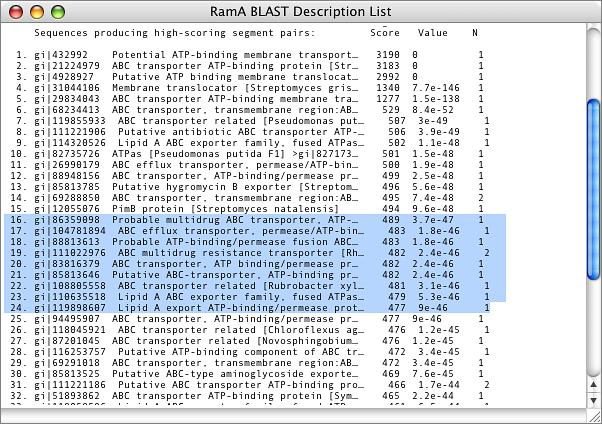
The Basic Local Alignment Search Tool (BLAST) algorithm is a popular program for identifying sequences in a databases that share sequence similarity to a search sequence. MacVector provides an integrated Internet BLAST interface to the databases at the NCBI. You can search using any of the blastn, blastp, tblastn, blastx and tblastx algorithms using any sequence databases available at the NCBI. The BLAST searches take place in the background, so you can continue to work with MacVector while one or more searches are in progress. As well as providing a standard BLAST report of the best aligning sequences in the database, MacVector also lets you select sequence in the “hitlist” and directly download those to your desktop or hard drive for further analysis.
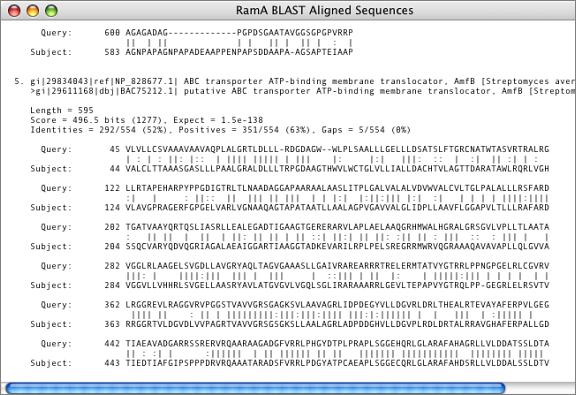
Align To Folder
As well as providing searches over the internet, MacVector has a built-in sequence searching facility that you can use to find similarities in collections of sequences stored on your own hard drive. The search algorithm is based on the popular FastA programs. You simply select a folder containing a collection of sequences and MacVector will scan through every sequence file in the folder looking for similarities. For this search, MacVector can use not only FastA formatted sequences, but also sequences in MacVector format, any chromatogram file format, and a number of simple text based formats.
As well as displaying a list of matching sequences and a graphical overview of the alignments, the Align to Folder function also generates a detailed text alignment showing each matching sequence aligned with the annotated search sequence. This is particularly useful for identifying the locations of sequence differences in relation to protein coding regions or other features on the search sequence.